NGS testing for Adventitious Agents
GMP-certified adventitious agent testing with NGS
Table Of Content
NGS testing for adventitious agent detection
NGS testing is suitable for broad detection of known and emerging viruses, using either a non-targeted or targeted sequencing approach. Non-targeted NGS can replace traditional in-vivo infectivity assays and supplement or replace in-vitro cell culture assays when demonstrated to be suitable for its intended purpose, without any head-to-head comparison. Traditional in-vitro and in-vivo infectivity assays rely on virus replication within specific systems for detection, limiting their ability to detect a wide range of viruses. Unlike these methods, NGS testing provides comprehensive detection and identification of both known and unknown viruses, without depending on virus growth or the specific biological characteristics required by traditional systems. NGS testing overcomes limitations of in-vitro AAT assays such as cell line susceptibility to infection, sample interference or cytotoxicity. ViruSure’s GMP-certified NGS testing can be used as an alternative or supplementing approach for traditional adventitious agent detection.
Testing for adventitious viruses is required for starting materials or intermediate products like cell banks, virus seeds, and unprocessed bulk harvests. According to ICH Q5A, 28-day testing on permissive cells is required for Master and Working Cell Banks (MCB/WCB) and Virus Seeds (MVS/WVS), with one subpassage after two weeks. For unprocessed bulk harvests, this approach may be reduced to 14 days with risk-based justification, considering factors like cell substrate, production duration, use of animal-derived materials, and viral clearance capacities.
High-Throughput Sequencing (HTS) can be used as an alternative to conventional methods for the detection of adventitious agents, especially for products where viral neutralization is not possible (e.g., viral vector vaccines, viral vector-derived products) or when sample toxicity interferes with traditional in-vitro or in-vivo assays. This approach offers a significant advantage for complex therapies such as cell and gene therapy (CGT) products, where QC testing often represents challenges due to limited sample volumes driven by high production costs and small batch sizes, typically associated with these products. In contrast to traditional testing methods, which require substantial sample volumes (approximately 23 mL per in-vivo test run or around 30 mL for in-vitro infectivity assays), NGS testing only needs around 200 µL for routine testing. This low sample volume requirements make NGS a more practical testing solution for CGT products.
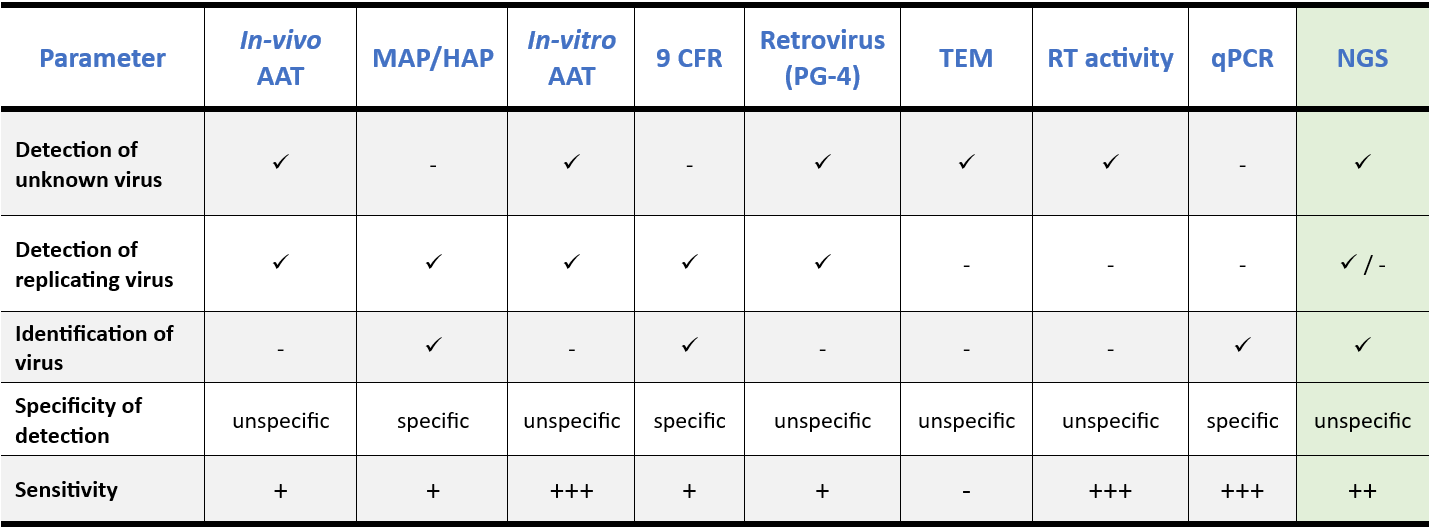
The table below highlights how NGS testing combines many advantages of traditional methods for adventitious agent detection into a single test. Compared to cell-based infectivity assays, in-vivo methods, and PCR, NGS testing offers a more efficient and comprehensive approach. Unlike PCR assays, NGS does not require prior knowledge of the contaminant’s genetic sequence, allowing for the detection of any virus present in a sample, including emerging ones, through sequence similarity searches against reference databases. By enabling simultaneous screening for all potential viral contaminants in a single run, NGS significantly reduces the time and effort required compared to running multiple PCR assays. Additionally, NGS testing eliminates the need for developing complex and costly neutralizing antibodies.
NGS validation and testing for adventitious agent detection
To ensure the effectiveness of Next-generation sequencing methods for applications such as cell bank, virus seed, or bulk harvest testing, a thorough validation is essential. As a qualitative limit test, NGS necessitates a comprehensive evaluation of specificity, the breadth of virus detection, and the limit of detection for each sample matrix. At ViruSure, NGS testing for adventitious agent detection employs a viromics approach, which sequences all viral nucleic acids encapsulated within intact viral capsids in a given sample. This method minimizes the risk of false positives by ensuring that only sequences protected by viral capsids are detected. Free DNA or RNA is eliminated by nuclease treatment, effectively reducing background interference from cellular nucleic acids.
Critical Steps in the NGS Workflow
Critical steps in the NGS workflow include: 1) sample pre-treatment and virus enrichment to enhance detection based on sample type; 2) efficient extraction of viral nucleic acids from both enveloped and non-enveloped particles; 3) appropriate library preparation for DNA and RNA viruses; 4) NGS sequencing run; and 5) bioinformatics analysis against a database containing diverse viral sequences. Follow-up strategies may be needed to confirm viral signals and distinguish them from non-viral sequences.
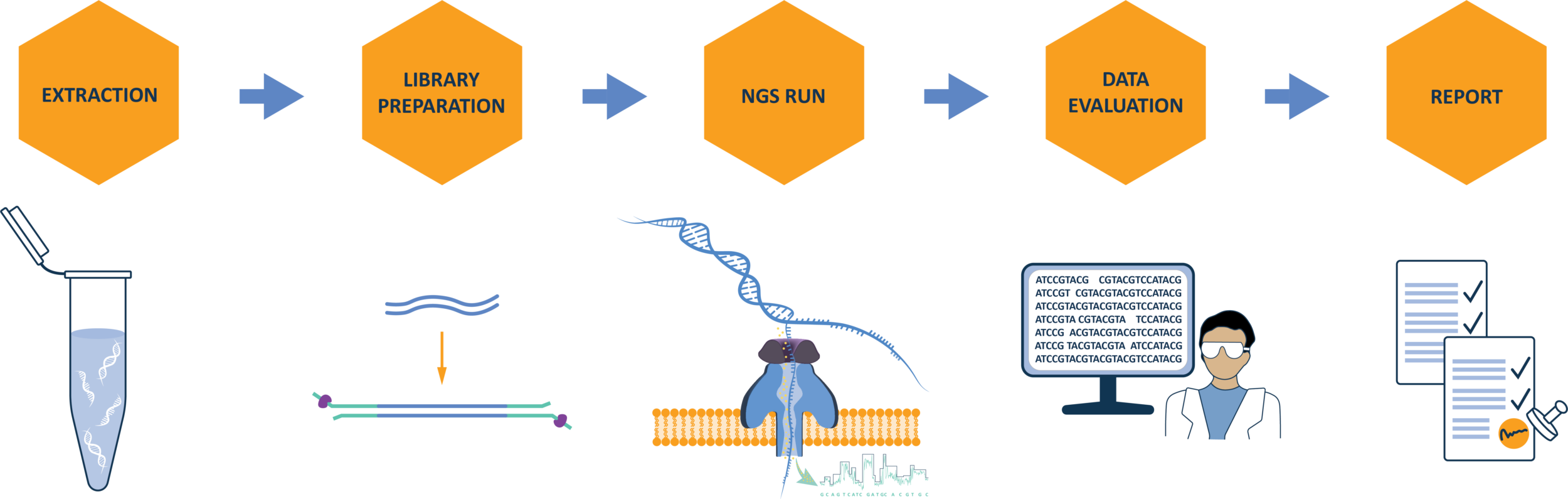
1) Sample pre-treatment:
Pre-treatments, such as nuclease digestion, centrifugation, or freeze-thaw cycles, improve NGS sensitivity by concentrating viral particles and reducing background cellular nucleic acids. These methods should be validated with model viruses in spiking studies to ensure their effectiveness.
2) Nucleic acid extraction:
RNA and DNA extraction can be done simultaneously or separately using different technologies (e.g. silica membranes, magnetic beads, or precipitation). The method should recover nucleic acids from all virus types and genomes, with an optional enrichment step to enhance sensitivity for low-level viral contamination.
3) Library preparation:
Library preparation ensures that the sample’s nucleic acid content can be sequenced on the chosen sequencing platform. For Nanopore´s long-read sequencing, this step involves ligation or tagmentation, where specific adaptors are attached to the ends of fragmented nucleic acids, enabling them to bind to the sequencing platform. Long-read sequencing platforms (e.g., PacBio, Oxford Nanopore) then sequence these extended fragments, facilitating the assembly of complete and continuous genomic sequences. Maintaining long nucleic acid fragments is key for long-read sequencing, as it enhances viral detection by the reduction of background noise obtained from random matches of sequencing reads against viral reference genomes. Barcoding (indexing) sequencing libraries helps identify cross-sample contamination.
4) NGS sequencing run:
During an NGS sequencing run, the number of reads generated influences the sensitivity for detecting adventitious viruses, as higher read numbers improve the likelihood of identifying low-level contaminants. To ensure reliable results, careful consideration must be given to factors such as sequencing depth, coverage, and contamination risks, particularly in multiplexed runs.
5) Bioinformatics analysis:
This final step involves building a pipeline to process the generated sequencing data. During this step, sequencing reads are compared with viral databases to identify a potential contaminant by sequence similarities. This can either be done with contigs generated from de-novo assembled reads or if long reads are used directly with the quality-controlled sequencing reads.
Proper method qualification and validation should utilize suitable reference materials, including virus panels with varying physical, chemical, and genomic characteristics, to demonstrate the workflow’s sensitivity, specificity, and detection range. A well-curated viral database encompassing diverse viral sequences is essential for broad virus detection. Additional types of reference materials can be employed to assess specific technical and bioinformatics steps.
Two-step NGS Validation Approach
NGS testing for adventitious agents
Routine sample testing with NGS
Upon completing matrix validation and establishing the limit of detection, the subsequent step is the testing of actual samples.
Reporting and bioinformatic analysis for virus detection
Our certificate of analysis for NGS adventitious agent testing, reviewed by our quality department to ensure full GMP compliance, provides a complete overview of the validation, testing, and bioinformatics analysis. It details the sensitivity and specificity of the assay for each sample matrix, alongside workflow descriptions, including sample preparation, sequencing, and bioinformatics analysis. Critical parameters, such as total reads, quality-control reads, and taxonomic classifications, are included, with clear criteria to differentiate true viral hits from false positives.
Our certificate of analysis provides transparent results, including an Excel summary of potential hits, with detailed rationale for their classification. With our extensive in-house expertise, we deliver high-quality, dependable results, making us a trusted partner in NGS adventitious agent testing.
All work adheres to GMP standards, meeting OECD guidelines, EU GMP directives, and US FDA 21 CFR Part 211.
Our bioinformatics pipeline combines decades of expertise and validated GMP-compliant workflows. If samples are multiplexed, bar-coded reads must first be demultiplexed, which is followed by the removal (trimming) of adapter sequences or low-quality regions and quality filtering. This initial quality control steps ensure that only high-quality data is used for subsequent analysis.
To enhance the efficiency and speed of the analysis, host or vector sequences may be subtracted. After the removal of host sequences, the remaining reads are mapped against viral and comprehensive databases to ensure accurate results and exclude false positive hits. Accurate results depend on well-annotated, comprehensive databases to reduce errors from misannotated or low-complexity sequences. Primary mapping of the data is performed against a complete viral database (e.g., RVDB) followed by counter-screening against a comprehensive nucleotide database (e.g., NCBI nt). Custom pipelines use nucleotide alignments to assess both known and unknown viruses. Tools, pipelines, and databases are systematically documented, and updates are qualified or revalidated to ensure accuracy and reliability.
Targeted and non-targeted NGS testing approaches
Our certificate of analysis for NGS adventitious agent testing, reviewed by our quality department to ensure full GMP compliance, provides a complete overview of the validation, testing, and bioinformatics analysis. It details the sensitivity and specificity of the assay for each sample matrix, alongside workflow descriptions, including sample preparation, sequencing, and bioinformatics analysis. Critical parameters, such as total reads, quality-control reads, and taxonomic classifications, are included, with clear criteria to differentiate true viral hits from false positives.
Our certificate of analysis provides transparent results, including an Excel summary of potential hits, with detailed rationale for their classification. With our extensive in-house expertise, we deliver high-quality, dependable results, making us a trusted partner in NGS adventitious agent testing.
All work adheres to GMP standards, meeting OECD guidelines, EU GMP directives, and US FDA 21 CFR Part 211.
Our bioinformatics pipeline combines decades of expertise and validated GMP-compliant workflows. If samples are multiplexed, bar-coded reads must first be demultiplexed, which is followed by the removal (trimming) of adapter sequences or low-quality regions and quality filtering. This initial quality control steps ensure that only high-quality data is used for subsequent analysis.
To enhance the efficiency and speed of the analysis, host or vector sequences may be subtracted. After the removal of host sequences, the remaining reads are mapped against viral and comprehensive databases to ensure accurate results and exclude false positive hits. Accurate results depend on well-annotated, comprehensive databases to reduce errors from misannotated or low-complexity sequences. Primary mapping of the data is performed against a complete viral database (e.g., RVDB) followed by counter-screening against a comprehensive nucleotide database (e.g., NCBI nt). Custom pipelines use nucleotide alignments to assess both known and unknown viruses. Tools, pipelines, and databases are systematically documented, and updates are qualified or revalidated to ensure accuracy and reliability.
What is the difference between genomics, transcriptomics, and viromics approach?
With NGS testing different approaches can be applied for comprehensive viral detection and characterization, including genomics (genomic viral nucleic acids), transcriptomics (viral mRNA), and viromics (encapsidated viral genomes). Each of these technologies is tailored to detect and analyze viral nucleic acids at different stages and forms within cell culture-derived materials:
1. Genomics involves the extraction of all nucleic acids from a sample, capturing both DNA and RNA to provide a complete profile. This approach is highly sensitive, allowing the detection of any viral genetic material present, ensuring that no potential signals are missed.
2. Transcriptomics focuses specifically on viral RNA within the sample, targeting RNA intermediates produced during viral replication. This method not only detects the presence of viruses but also suggests active viral replication, offering a more immediate indicator of viral activity.
3. Viromics approach specifically targets intact viral particles. Prior to viral genome extraction, nucleic acids from host cells and other free DNA/RNA are removed using nuclease treatment. This process ensures that only intact viral particles remain for subsequent extraction and sequencing, providing a strong indication that any detected signal may originate from viable, potentially infectious virus.
The application of NGS for adventitious virus detection offers enhanced sensitivity, capable of detecting a wide spectrum of viruses, including unknown or emerging ones, and is supported by regulatory guidelines. This advanced detection capability is critical for ensuring the safety of viral vector-based products (e.g., AAVs, viral vaccines, gene therapy vectors) and cell culture-derived therapies, overcoming limitations associated with traditional assays and enabling a more comprehensive assessment of viral presence in complex sample matrices.
If you’re interested in learning more, don’t hesitate to reach out
